Your reference partner for
What's in it for me ? Digital pathology for :
Discover Cytomine
We import your microscope and your image-analysis software in your web browser to help you improve education, science and diagnosis in health and biomedical fields.
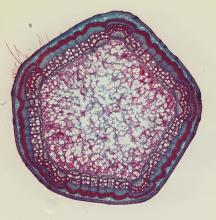
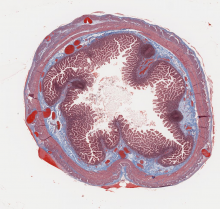
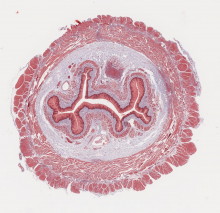
open access image collection
Here are 3 examples. Access our collection and explore all the images inside Cytomine.
Testimonials

Collaborative biomedical image analysis
Installation, maintenance and support of Cytomine.
On-demand algorithm development.

Open source image analysis
Open source internet application for collaborative analysis of multi-gigapixel images.
Our network